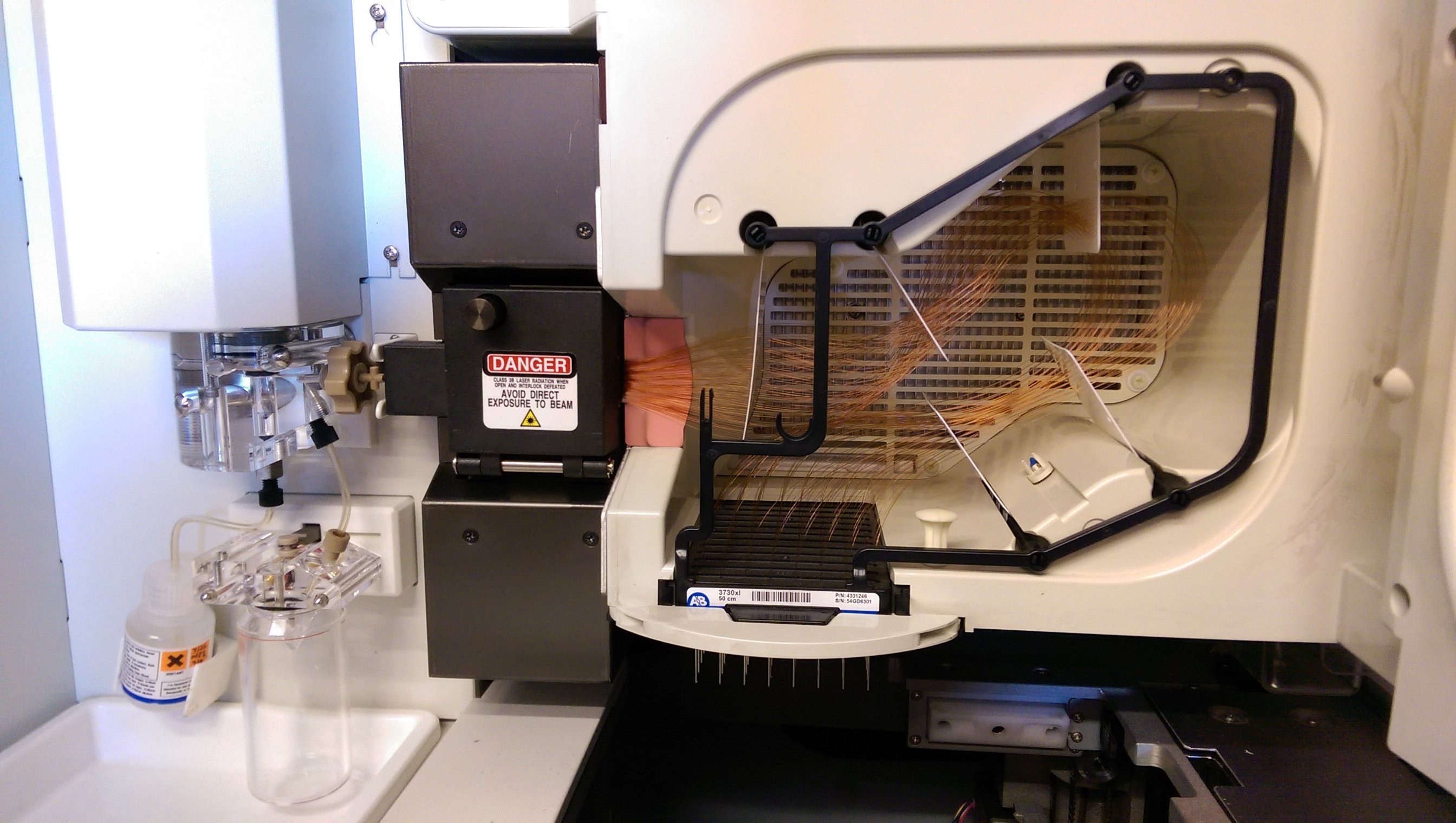
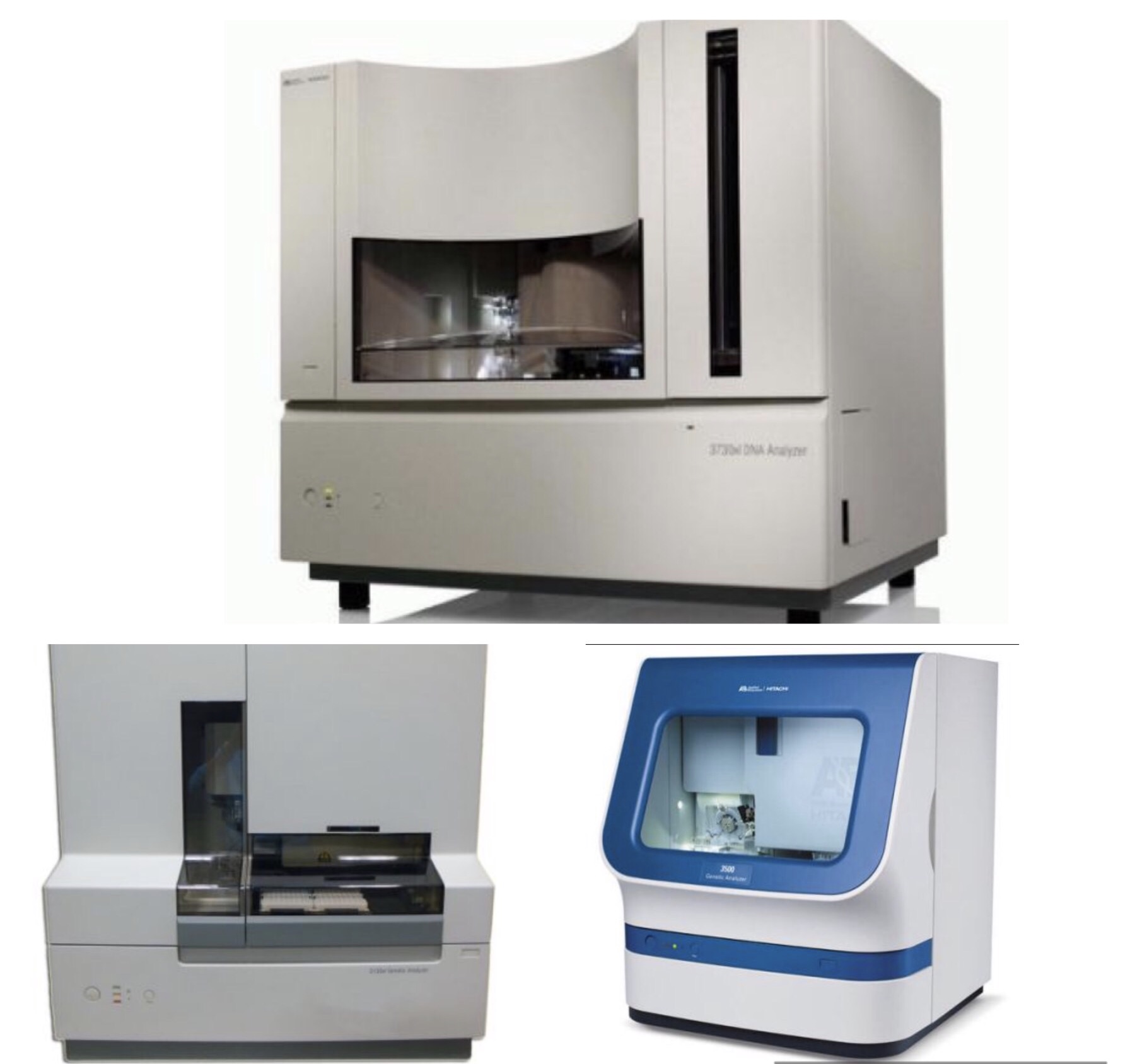
One of the most common complaints we hear from our customers is they are seeing less than optimal results in their sequencing data. After making sure your machine is working properly, we often come to the conclusion that the issue is related to the capillary array.
Most of the time, the problem is simply a dirty capillary array. Over time, dried and degraded polymer will accumulate on the inner walls of your capillary array. This can adversely affect the resolution during sample runs and cause your results to suffer. Some common symptoms of a failing array are:
- Loss of resolution
- Broadening or “fat” peaks
- Delayed DNA peaks
- Blue or yellow bands in some, but not all of the capillaries
- One or two capillaries showing no peaks at all



.jpg)